cpRNPs guarantee global chloroplast RNA stability
 Chloroplast mRNAs must be acted upon by numerous factors before they can serve as templates for the translational apparatus. Various factors required for the cleavage, splicing and editing of specific mRNAs have been described in recent years. Not surprisingly, many of these factors are RNA-binding proteins (RBPs). In contrast to the successful efforts of various laboratories to identify and characterize RBPs in the chloroplast, relatively little progress has been made towards understanding:
Chloroplast mRNAs must be acted upon by numerous factors before they can serve as templates for the translational apparatus. Various factors required for the cleavage, splicing and editing of specific mRNAs have been described in recent years. Not surprisingly, many of these factors are RNA-binding proteins (RBPs). In contrast to the successful efforts of various laboratories to identify and characterize RBPs in the chloroplast, relatively little progress has been made towards understanding:
(i) the functionally relevant mechanistic details of chloroplast protein–RNA interactions; and
(ii) the regulatory roles of these RBPs in chloroplast biogenesis.
It is not yet known how the chloroplast RBPs fulfill their functions on a molecular level, or how they impact the production of chloroplast proteins. Most chloroplast RBPs, such as most pentatricopeptide repeat (PPR) proteins, are part of the constitutive expression machinery of the chloroplast, and thus seem unlikely to perform the short-term regulatory tasks involved in adapting the chloroplast to external or internal stimuli. In contrast, chloroplast RNA-recognition motif (RRM) proteins respond to various signals and appear to be prime candidates for regulating chloroplast gene expression.
The nuclear–encoded chloroplast ribonucleoproteins (cpRNPs) are a group of plastid hnRNP-like RNA-binding proteins that have two RRM motifs. They are highly abundant proteins that are regulated by various environmental stimuli. cpRNPs bind multiple RNA targets and are involved in RNA processing, stabilization and editing.
CP31A
The Arabidopsis cpRNP CP31A contains two RNA recognition motifs (RRMs) and an unstructured acidic domain that can be phosphorylated. CP31A was shown to be important for RNA editing and the stabilization of certain mRNAs, especially the ndhF transcript. Furthermore, null mutants of CP31A show bleaching of newly emerging tissue and a global reduction in mRNA accumulation at low temperatures.
Expression of a CP31A version without its acidic domain (CP31A?AD) in cp31a null mutants rescued the loss of the ndhF transcript as well as the RNA editing defects under normal growth conditions. However, bleaching of the newly emerging tissue in the cold was not complemented. We hypothesized that CP31A shows different patterns of transcript association under normal and cold conditions with the acidic domain playing a prominent role in regulated RNA interactions. To test this, RNA immunoprecipitation and deep sequencing (RIP-seq) from full-length and AD-less CP31A was performed. This analysis demonstrates that CP31A shows increased binding to RNA in the cold and that its acidic domain is supportive, but not essential for cold-induced RNA binding.

CP33A
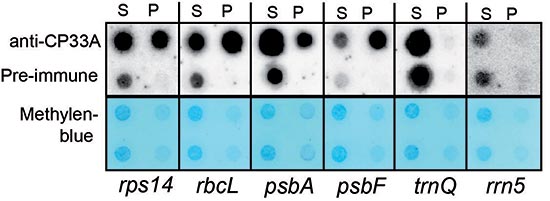
We have shown that the cpRNP CP33A localizes to a distinct sub-chloroplastic domain and is essential for chloroplast development. The loss of CP33A yields albino seedlings that exhibit aberrant leaf development and can only survive in the presence of an external carbon source. Genome-wide RNA association studies demonstrate that CP33A associates with all chloroplast mRNAs. For a given transcript, quantification of CP33A-bound versus free RNAs demonstrates that CP33A associates with the majority of most mRNAs analyzed. Our results further show that CP33A is required for the accumulation of a number of tested mRNAs, and is particularly relevant for unspliced and unprocessed precursor mRNAs. Finally, CP33A fails to associate with polysomes or to strongly co-precipitate with ribosomal RNA, suggesting that it defines a ribodomain that is separate from the chloroplast translation machinery. Collectively, these findings suggest that CP33A contributes to globally essential RNA processes in the chloroplasts of higher plants.